Review of B Cells Cd4+ T Cells and Cd8+ T Cells
Dendritic Cells as Antigen Presenting Cells to Initiate a Principal Response
Initiation of an adaptive immune response begins with naïve T cells being activated past antigens presented on dendritic cells (DCs), a highly specialized professional antigen presenting cell (APC) (1, 2). As a frontline defender, DCs are primal APCs bridging the gap between innate and adaptive amnesty. Located primarily in peripheral tissues, immature DCs are well known for their power to recognize and capture invading pathogens mainly through phagocytosis and micropinocytosis. Uptake of antigen (Ag) is closely followed by upregulation of MHC Course I, Class Ii and co-stimulatory molecules on the surface of DCs, equally they lose the power to perform macropinocytosis (iii, 4). Mature DCs so migrate to draining lymph nodes where they present pathogen-derived epitopes to naïve CD4 and CD8 T cells (5). 1 unique characteristic of DCs is their ability to uptake Ag via phagocytosis and cantankerous-present it on MHC Class I molecules. This makes DCs a perfect principal antigen presenter for initiation of an immune response (2). Moreover, it has also been noted that activation of immature DCs by various Toll-like receptor ligands (TLR3 and TLR9) transiently increases antigen specific micropinocytosis (half dozen), which probable increases the ability of DCs to capture Ag within an inflammatory context. To appointment, inquiry into what contributions DCs make to retentiveness development has been limited and mainly focuses on memory CD8 T cells evolution (7–11). Apply of Batf3 knock-out (KO) mice, which lack CD8a DCs responsible for cantankerous-presentation, found no touch on on principal CD4 T cell responses but drastically impaired CD8 responses (12). Likewise, work using Toxoplasma gondii showed a crucial part for CD4 T cells in protecting Batf3 KO mice from succumbing to T. gondii infection (xiii). Yet none of these studies using DC KO mice investigated a role for DCs in memory CD4 T prison cell development. Information from Dalai et al., however suggests that loss of DCs does not likely impact the germination of memory CD4 T cells as removal of CD11c+ DCs did non affect the development of quiescent retentivity CD4 T cells (14).
B Cells equally APCs in Secondary Responses
B cells are another major professional APCs, which unlike DCs, accept upward antigens specifically by B cell receptor (BCR) (1). Upon interaction with a cognate Ag, the BCR-Ag circuitous would be internalized and shuttled to the specialized MHC form II enriched compartments (MIIC) for processing and presentation to the Ag-specific CD4 T cells (fifteen). These CD4 T-B interactions provide essential activation signals to B cells for affinity maturation and differentiation into retentiveness B, or antibiotic-secreting plasma B cells (xvi). The retentiveness B cells generated from this T-B interaction have been found to also be important for CD4 T cell memory responses (17).
How B Cells and DCs Impact Memory T Jail cell Evolution
It is generally accustomed that retentivity T cells differentiate afterwards exposure to Ag followed by multiple rounds of proliferation (18–twenty). While characterization of retentiveness T cells has been explored intensely, the onset of differentiation of Ag-experienced T cells into memory, and how APCs influence this process is less appreciated. Especially that in rare publications, it has been proposed that CD8 retentivity T cells may be generated upon asymmetric prison cell partitioning, which precludes the demand for interaction with antigen presenting cells (14, 21, 22). On another line of studies, CD8 retention T cell evolution and homeostasis has been reported to be mediated by IL-15Rα expressed by DCs and Macrophages (23, 24). It is too constitute that long-lasting CD8 memory can be achieved in the absence of CD4 T cells or B cells (25).
For CD4 Memory T evolution, withal, TCR-pMHC interaction appears to bulldoze CD4 Memory T development (14, 26–30). In this regard, Williams et al. found that lower levels of LCMV antigen density led to high functional ardor CD4 T retentiveness differentiation, while college levels of LCMV antigen density promoted both loftier avidity and low ardor CD4 T cells expansion (28). Still, the authors did non explore whether DCs or B cells were the APCs to drive such differentiation. Studies addressing contributions of B cells to activation of naïve CD4 T cells has been inconclusive (31). Conversely, several investigations have reported that B cells play a critical function in regulating CD4 memory T development and differentiation (14, 17, 26, xxx, 32–37). It is noteworthy that among these studies, both Chowdhury (17) and Misumi (35) establish that absence of antigen specific B cells either from SCID mice without B cells or treatment of anti-CD20 mAb did non touch the priming of CD4 T cells in viral infection but impaired the development and effector part of memory CD4 T cells. By virtue of having antigen specific B Cell Receptors, B cells can recognize and internalize specific antigens, process, and present them to cognate CD4 T cells (fifteen). Equally such, B cell antigen presentation adds a new and exciting dimension to our current cognition.
The first clear demonstration that B cells play a role in memory CD4 T cell generation/differentiation came from Bradley and colleagues who reported B prison cell knockout mice did not develop memory CD4 T cells (32). Further studies take shown that loss of B cells adversely affects development of Tuberculosis (TB)-specific CD4 retentivity precursor effector cells (MPECs) in TB vaccinated B cell scarce mice (36). Because of the ability of B cells to produce antibodies that bind to Ag, it has been postulated that contribution of B cells to CD4 retentivity T cell development might exist linked to Ag-Ab complexes. However, when this issue was specifically addressed by Whitmire et al., T cell responses to lymphocytic choriomeningitis virus (LCMV) infection, the squad found that in contrast to B prison cell-deficient mice, membrane Ig expressing Tg mice retained functional Th jail cell memory, indicating that B cells selectively preserve CD4 T prison cell memory independently of allowed complex formation (33).
To direct test if B cells were important for the development of CD4 T jail cell retention, Dalai et al. tested the specific interactions of various APCs with Ag experienced CD4 T cells (14). Using an ex vivo anergy analysis, the group showed that but B cells, but non DCs, induced a resting state in Ag experienced CD4 T cells. Farther in vivo label using an adoptive prison cell transfer assay further confirmed the ex vivo observations. Previous findings had demonstrated that sub-optimal levels of agonist peptides had induced a resting state in T cells in vitro, and in vivo (34, 38–44). Thus, the above observations that B cells, simply not DCs, pulsed with low doses of Ag induced resting retentiveness CD4 T cells confirmed prior findings that B cells are indispensable for retention CD4 T cell development/differentiation. In agreement with the above findings, B cell deficient mice did not develop quiescent CD4 memory T cells. However, when B cells were transferred to the B jail cell scarce mice, hyporesponsive CD4 memory T cells were developed. Chiefly, B2 (B220+CD43+) follicular B cells, which take diverse BCR were identified equally the cells that rendered CD4 retentiveness T cells hyporesponsive (fourteen). These finding were afterward supported by Keck et al, who found that B cells were required for both optimal expansion and T-bet expression in response to weak TCR stimulation and optimal generation of CD4 T memory (30).
Contribution of Ag Density Presented by Follicular B Cells to CD4 Retentivity T Cell Consecration/Differentiation
Building upon those initial findings, Dalai et al. tested the effects of B cell presentation of peptide-MHC (pMHC) density on the induction of quiescent retentiveness CD4 T cells. They used a clever strategy past recovering B cells from mice at various timepoints post immunization and transferring them into recipient mice harboring CD4 T retentiveness forerunner cells at 4-day intervals (26). This staggered timeframe immune Dalai et al. to correlate the amount of pMHC presented by the B cells to the fourth dimension post immunization; earlier time points displayed more pMHC, and afterwards time points fewer pMHC. Interestingly, the group found that but B cells harvested between twenty-four hours 16-20 post OVA immunization induced resting hyporesponsive CD4 retentiveness T cells. These findings supported the idea that CD4 memory T cells are signaled to a resting state past the presentation of a subthreshold numbers of pMHC. These conclusions were further expanded to HEL-specific B cells (45) HEL-specific B cells when used for induction of quiescence/resting state of Ag experienced T cells were more efficient in capturing the Ag and induced quiescence in Ag experienced CD4 T cells at much after time points, i.e., 41-48 days vs 16-twenty days post immunization past non-specific B cells. In those experiments B cells immunized with poly peptide antigens were transferred to mice that carried primed T cells at 4-mean solar day intervals. The rationale was to detect out when during an immune response B cell presentation of pMHC reaches to the levels necessary for the induction of quiescence naturally, in vivo. It was quite gratifying to come across that HEL-specific B cells had captured far more than antigen so that the required densities of pMHC for inducing quiescence had reached 20 plus days after than the polyclonal B cells (26). Altogether, Dalai et al. established that: (i) B cells are the APCs responsible for rendering CD4 retentivity T cells the quiescent, and (two) depression levels of pMHC presentation are the master driving force that signal CD4 T cells to enter a resting country (26).
More than recently, we accept explored how this country of anergy impacts both the longevity and function of CD4 T memory cells. Song et al. investigated cistron expression dynamics in CD4 T retentivity cells at different stages postal service immunization representing activated, early on memory, late memory, and long-term memory stages (46). OVA-specific DO11.10 T cells were adoptively transferred into naïve mice before infecting them with Vaccinia-OVA virus, followed by harvesting the CD44hiDO11.onepos T cells at unlike time points postal service immunization and subjecting their mRNA for gene expression analyses. Through this approach, the group was able to illustrate the factor expression dynamics occurring during CD4 T memory development up to nigh 1 year. In agreement with findings of others (47–51), authors found that the OVA-specific CD4 memory T cells adopted a resting phenotype. Furthermore, the memory phenotype associated with multiple genetic programs regulating cellular proliferation, DNA repair, prevention of apoptosis, glucose, and lipid metabolism (Effigy one). Specifically, most genes regulating cellular proliferation and DNA repair response were found to exist associated with p53 pathways, which highlights the importance of limiting cell proliferation and promoting DNA repair in long-lived CD4 Memory T cells. As well, of note was that similar CD8 Memory T cells, genes regulating lipid metabolism were upregulated indicating that long-lived CD4 Memory T cells may also rely on lipid metabolism. However, unlike CD8 memory, the genes regulating lipid metabolism in CD4 T retention were plant to exist centered on regulating cellular cholesterol and ceramide levels, which could be related to the T cell signaling and prevention of apoptosis. Altogether, these programs play important roles in CD4 Memory T development and maintenance.
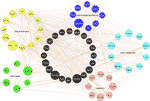
Effigy one Cistron networks in long-lived CD4 memory T cells. Five different factor programs were identified equally dynamically regulated during memory CD4 T cell differentiation. The genes shown were from the long-lived memory CD4 cells 10.5 months post immunization as compared to naïve controls and are marked in different colors: Xanthous: cell proliferation; Dark-green: Dna repair; Red: Apoptosis; Light blueish: Lipid metabolism; Dark bluish: Carbohydrate metabolism; Black: Not identified in the five gene programs simply served as connecting genes. Each line represents an interaction/co-expression of genes as identified by literature report.
The above genetic studies as well revealed upregulated levels of CD99, CCR10 and Itga3 as potentially new surface markers for long-lived CD4 memory T cells. Importantly, the high expression levels of these new CD4 retention markers at the poly peptide level were confirmed to hold truthful across unlike animal models and antigens. For example, CD99hi resting human CD4 T cells from flu vaccinated donors had much ameliorate proliferation responses than the CD99lo CD4 subsets to in vitro challenges, indicating that the gene expression programs plant in murine CD4 memory T cells could also be applicable to human CD4 memory T cells. Overall speaking, this work indicated not only that the resting state of CD4 memory T cells was mediated by multiple genes and could be office of the reason for CD4 memory longevity, but also the surprising findings that the murine CD4 memory differentiation is regulated by genetic programs that evolve upwardly of half dozen months to fully appear.
Contribution of Form II Accessory Molecules in CD4 Memory Formation
The finding that proper evolution of CD4 T memory cells relies on quantitative differences in presentation of immunodominant epitopes by B cells, brings the focus to the potential roles that accessory molecules in antigen processing play in the selection of epitopes for binding to MHC Course Two. It is demonstrated that equally the primary Form 2 peptide-editor, HLA-DM (human DM; murine H2-M) contributes to the option of immunodominant epitopes by generating higher quantities of those epitopes (52–55). HLA-DO (human being Exercise; murine H2-O), is a 2nd accompaniment molecule, which requires DM for its expression; Practice is mainly expressed in thymic epithelium and B cells (54–56). Both DM and DO contribute to T cell amnesty in a significant fashion, because lymphocytes normally respond to a small portion of the potential determinants on a poly peptide antigen, defined as 'immunodominant' (57). Immunodominant epitopes are the essential targets of the allowed response confronting infectious diseases, cancer, autoimmune diseases, and allergy. Hence, deserve the attention devoted to the agreement of epitope selection and immunodominance. To improve sympathise how each accessory molecule impacts immunodominant epitope choice, we must hash out each molecule individually.
Mechanism of DM in Finding the Immunodominant Determinants During Antigen Processing
Information technology has been well established that the MHC Two groove is flexible and requires a leap peptide to maintain its shape. Without a peptide, the MHC II groove would close and becomes inefficient in binding peptides (58–60). Thus, newly synthesized MHC 2 molecules demark to a domain of the Class II invariant chain (Prune) that serves ii functions; a) protects the groove from bounden to peptide in the ER (61), and b) acts as a place-keeper, while some other domain of Ii guides the complex to the specialized vesicular compartments filled with pathogen-derived antigenic peptides, MIIC. Inside MIIC, DM is necessary to showtime dissociate Clip to form a peptide-receptive conformation that can quickly scan unfolded exogenous proteins to notice its suitable determinant (62). DM does this chore past finer dissociating whatsoever peptide sequences that do not make full in the pockets of the MHC Two groove. Only when a sequence of antigenic determinant that would fit in the MHC Ii groove leading to formation of a compact folded conformation, the circuitous becomes resistant to DM-mediated dissociation (DM-resistant). Next, the proteases would trim the MHC II bound determinant. The proteases also cut the antigenic determinants that practice not fit the groove, hence are susceptible to DM-mediated dissociation (DM-sensitive) and are dislodged by DM (63–71). The solution of the crystal structure of the DM/DR complex (72) using DR1/peptide complexes that enforced an open up DR1 groove, revealed that DM would bind the P1 pocket of HLA-DR molecules tightly if empty, and would remain jump until a P1 filling peptide would bind the groove and induce endmost of the groove, and displacing DM (72–74). The to a higher place findings were complemented by the measured thermodynamics of peptide binding to DR1, indicating that a greater entropic penalty, versus a smaller penalty, was associated with structural rigidity rather than with the flexibility of the pMHC complexes (75). These findings suggested that an overall dynamic MHC II conformation in improver to P1 pocket occupancy, determines susceptibility to DM-mediated peptide substitution and provides a molecular mechanism for DM to efficiently target poorly fitting pMHC II complexes and editing them for more stable ones. Hence, in addition to the removal of CLIP, DM helps in shaping epitope option and immunodominance by producing a higher affluence of those determinants (62).
Dissimilar Models on How Practice Fine-Tunes Antigenic Epitope Selection
Exercise also contributes to the selection of immunodominant epitopes, although agreement the contributions of DO to epitope selection has proven to exist highly challenging (54–56, 76). In brief, our cognition well-nigh DO can be distilled into two working hypotheses: (1) DO binds to DM to inhibit its action, mainly removal of the Clip peptide and, (2) DO differentially affects presentation of structurally diverse peptides and acts equally a 2d accompaniment molecule working together with DM in fine tuning MHC II repertoire selection. Data in support of the one-time hypothesis mainly comes from studying over-expression of Practise genes in cell lines, or dendritic cells (77, 78); Welsh, 2019 #13} and the recent mutagenesis and structural studies of DM/DO interactions (79, 80). The 3D structure of DM/DO showed that DO binds to DM at the same interface with which DM interacts with DR1 (74). Studies supporting the latter hypothesis came from biochemical (81) and biophysical studies demonstrating that DO only affected association kinetics of sure peptides to DR but, had no outcome on the dissociation kinetics of any tested peptide/DR1 complexes (76, 82). The effects of DO on association kinetics straight correlated with peptide sensitivity to DM-mediated dissociation. DO reduced binding of peptides that formed DM-sensitive complexes with DR and enhanced the bounden of peptides that formed DM-resistant complexes. In a nutshell, it was conspicuously shown that; i) DO works directly on DR1, and not by regulating the upshot of DM, ii) Exercise tin only bind the peptide-receptive MHC Grade II, and iii) that this peptide-receptive conformation is generated past DM. Hence, authors proposed that DM and DO cooperate for a more than effective epitope selection. Thus, in i model, Do would reduce presentation of immunodominant epitopes, whereas in the other, DO would increase the abundance of immunodominant epitopes.
Speculations for Future Research
The question of the potential contributions of DO and DM to memory CD4 T prison cell evolution is of most interest and is discussed below. A few characteristics of DO hint to its possible link to CD4 memory differentiation. First, DO is mainly expressed in B cells (81, 83, 84) and it enhances the presentation of immunodominant epitopes (56, 76). Side by side, it has been documented that successful entry of B cells into the germinal center (GC) requires loftier expression levels of pMHC (85–89). B cells enter GC and interact with CD4 T cells in search of proper signaling for affinity maturation. Information technology is conceivable that CD4 T cells as well receive signals from GC B cells for their ain differentiation into resting retentivity T cells. One might say if high levels of pMHC equip B cells for entry into GC, how could B cells signal T cells to differentiate into resting retentiveness, as this procedure requires suboptimal densities of pMHC presentation. An answer worth considering is that once B cells enter GC, their expression levels of DO and DM decreases, leading to a reduced level of pMHC II expression (90–92). As such, those GC B cells tin interact with Ag-specific CD4 T cells in the Light Zone (LZ), selecting them to become memory forerunner cells. In support of this argument, in an elegant written report, Kim et al. accept documented that memory CD4 T cells bear high affinity TCR for pMHC II (27), hence memory CD4 T cells are selected based on TCR affinity. One may predict that alterations in this controlled entry into the GC reaction could lead to faulty CD4 T cell memory development and possibly the evolution of increased autoreactivity.
Since biology tends to repeat itself, it would be interesting to compare the effects of pMHC numbers on APCs and their effects on CD8 retentivity T cell development. While equally far equally we know no studies has made such data available, in an heady new study authors reported that in the absence of B cells CD8 T Cell memory germination was compromised, while CD8 effector role was enhanced. 1 might speculate that since CD4 T cells are essential for CD8 memory T jail cell evolution (93), perhaps their contributions to CD8 retention is mediated indirectly via CD4 retentiveness T cells.
Hereafter experimental bear witness is needed to clarify the proposed relationship of these MHC II accessory molecules to the development and maintenance of CD4 memory T cells, and hopefully this review would prompt new research on the qualitative and quantitative antigen presentation on CD8 retention T cell development.
Author Contributions
All authors listed have fabricated a substantial, directly, and intellectual contribution to the work and approved information technology for publication.
Funding
Supported past grants from NIAID, R01AI063764, R21AI101987, and R01AI120634, to SS-N.
Disharmonize of Interest
The authors declare that the research was conducted in the absence of whatsoever commercial or fiscal relationships that could be construed equally a potential conflict of interest.
References
1. Tater Grand, Weaver C. Janeway'south Immunobiology. New York & London: Garland Science (2016).
Google Scholar
2. Itano AA, McSorley SJ, Reinhardt RL, Ehst BD, Ingulli E, Rudensky AY, et al. Distinct Dendritic Cell Populations Sequentially Nowadays Antigen to CD4 T Cells and Stimulate Dissimilar Aspects of Cell-Mediated Immunity. Immunity (2003) 19:47–57. doi: 10.1016/S1074-7613(03)00175-four
PubMed Abstract | CrossRef Full Text | Google Scholar
3. Sallusto F, Cella M, Danieli C, Lanzavecchia A. Dendritic Cells Utilise Macropinocytosis and the Mannose Receptor to Concentrate Macromolecules in the Major Histocompatibility Circuitous Class 2 Compartment: Downregulation by Cytokines and Bacterial Products. J Exp Med (1995) 182:389–400. doi: x.1084/jem.182.ii.389
PubMed Abstract | CrossRef Full Text | Google Scholar
iv. Lutz MB, Schuler One thousand. Immature, Semi-Mature and Fully Mature Dendritic Cells: Which Signals Induce Tolerance or Immunity? Trends Immunol (2002) 23:445–9. doi: 10.1016/S1471-4906(02)02281-0
PubMed Abstruse | CrossRef Full Text | Google Scholar
6. West MA, Wallin RP, Matthews SP, Svensson HG, Zaru R, Ljunggren HG, et al. Enhanced Dendritic Jail cell Antigen Capture Via Toll-Similar Receptor-Induced Actin Remodeling. Scientific discipline (2004) 305:1153–vii. doi: 10.1126/science.1099153
PubMed Abstruse | CrossRef Full Text | Google Scholar
7. Zammit DJ, Lefrancois L. Dendritic Prison cell-T Cell Interactions in the Generation and Maintenance of CD8 T Prison cell Memory. Microbes Infect (2006) 8:1108–15. doi: 10.1016/j.micinf.2005.12.002
PubMed Abstract | CrossRef Full Text | Google Scholar
9. Enamorado M, Khouili SC, Iborra S, Sancho D. Genealogy, Dendritic Jail cell Priming, and Differentiation of Tissue-Resident Memory Cd8(+) T Cells. Front Immunol (2018) nine:1751. doi: 10.3389/fimmu.2018.01751
PubMed Abstract | CrossRef Full Text | Google Scholar
10. Yu B, Zhang K, Milner JJ, Toma C, Chen R, Scott-Browne JP, et al. Epigenetic Landscapes Reveal Transcription Factors That Regulate CD8(+) T Cell Differentiation. Nat Immunol (2017) 18:573–82. doi: 10.1038/ni.3706
PubMed Abstruse | CrossRef Full Text | Google Scholar
11. Johnnidis JB, Muroyama Y, Ngiow SF, Chen Z, Manne S, Cai Z, et al. Inhibitory Signaling Sustains a Distinct Early Retention CD8(+) T Prison cell Precursor That Is Resistant to DNA Impairment. Sci Immunol (2021) half dozen:one–16. doi: 10.1126/sciimmunol.abe3702
CrossRef Total Text | Google Scholar
12. Hildner Chiliad, Edelson BT, Purtha WE, Diamond G, Matsushita H, Kohyama Grand, et al. Batf3 Deficiency Reveals a Disquisitional Role for CD8alpha+ Dendritic Cells in Cytotoxic T Prison cell Immunity. Science (2008) 322:1097–100. doi: 10.1126/scientific discipline.1164206
PubMed Abstract | CrossRef Full Text | Google Scholar
thirteen. Tussiwand R, Behnke MS, Kretzer NM, Grajales-Reyes GE, Murphy TL, Schreiber RD, et al. An Important Role for CD4(+) T Cells in Adaptive Amnesty to Toxoplasma Gondii in Mice Lacking the Transcription Factor Batf3. mSphere (2020) 5:1–11. doi: 10.1128/mSphere.00634-20
CrossRef Full Text | Google Scholar
fifteen. Adler LN, Jiang W, Bhamidipati K, Millican M, Macaubas C, Hung SC, et al. The Other Office: Class 2-Restricted Antigen Presentation by B Cells. Front Immunol (2017) 8:319. doi: 10.3389/fimmu.2017.00319
PubMed Abstract | CrossRef Full Text | Google Scholar
17. Chowdhury MG, Maeda M, Yasutomo K, Maekawa Y, Furukawa A, Azuma M, et al. Antigen-Specific B Cells Are Required for the Secondary Response of T Cells But Not for Their Priming. Eur J Immunol (1996) 26:1628–33. doi: 10.1002/eji.1830260733
PubMed Abstract | CrossRef Full Text | Google Scholar
19. van Stipdonk MJ, Lemmens EE, Schoenberger SP. Naive CTLs Require a Single Cursory Period of Antigenic Stimulation for Clonal Expansion and Differentiation. Nat Immunol (2001) 2:423–ix. doi: 10.1038/87730
PubMed Abstract | CrossRef Full Text | Google Scholar
21. Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, et al. Disproportionate T Lymphocyte Division in the Initiation of Adaptive Immune Responses. Science (2007) 315:1687–91. doi: 10.1126/science.1139393
PubMed Abstruse | CrossRef Full Text | Google Scholar
22. Pollizzi KN, Sun IH, Patel CH, Lo YC, Oh MH, Waickman AT, et al. Asymmetric Inheritance of mTORC1 Kinase Activity During Segmentation Dictates CD8(+) T Cell Differentiation. Nat Immunol (2016) 17:704–11. doi: 10.1038/ni.3438
PubMed Abstract | CrossRef Total Text | Google Scholar
23. Stonier SW, Ma LJ, Castillo EF, Schluns KS. Dendritic Cells Drive Retentivity CD8 T-Cell Homeostasis Via IL-15 Transpresentation. Blood (2008) 112:4546–54. doi: ten.1182/blood-2008-05-156307
PubMed Abstract | CrossRef Full Text | Google Scholar
24. Mortier E, Advincula R, Kim L, Chmura S, Barrera J, Reizis B, et al. Macrophage- and Dendritic-Cell-Derived Interleukin-xv Receptor Blastoff Supports Homeostasis of Distinct CD8+ T Prison cell Subsets. Immunity (2009) 31:811–22. doi: 10.1016/j.immuni.2009.09.017
PubMed Abstract | CrossRef Full Text | Google Scholar
25. Di Rosa F, Matzinger P. Long-Lasting CD8 T Cell Memory in the Absenteeism of CD4 T Cells or B Cells. J Exp Med (1996) 183:2153–63.
PubMed Abstract | Google Scholar
26. Dalai SK, Khoruzhenko Due south, Drake CG, Jie CC, Sadegh-Nasseri S. Resolution o Infection Promotes a State of Dormancy and Long Survival of CD4 Retention T Cells. Immunol Cell Biol (2011) 89(8):870–81. doi: 10.1038/icb.2011.2
PubMed Abstract | CrossRef Full Text | Google Scholar
27. Kim C, Wilson T, Fischer KF, Williams MA. Sustained Interactions Between T Jail cell Receptors and Antigens Promote the Differentiation of CD4(+) Memory T Cells. Amnesty (2013) 39:508–20. doi: 10.1016/j.immuni.2013.08.033
PubMed Abstruse | CrossRef Full Text | Google Scholar
28. Williams MA, Ravkov EV, Bevan MJ. Rapid Culling of the CD4+ T Cell Repertoire in the Transition From Effector to Memory. Immunity (2008) 28:533–45. doi: ten.1016/j.immuni.2008.02.014
PubMed Abstract | CrossRef Full Text | Google Scholar
29. Rees W, Bender J, Teague TK, Kedl RM, Crawford F, Marrack P, et al. An Inverse Relationship Between T Cell Receptor Analogousness and Antigen Dose During CD4(+) T Prison cell Responses In Vivo and In Vitro. Proc Natl Acad Sci United states of america (1999) 96:9781–half dozen. doi: 10.1073/pnas.96.17.9781
PubMed Abstruse | CrossRef Full Text | Google Scholar
30. Keck Due south, Schmaler Yard, Ganter Southward, Wyss L, Oberle S, Huseby ES, et al. Antigen Affinity and Antigen Dose Exert Distinct Influences on CD4 T-Cell Differentiation. Proc Natl Acad Sci Usa (2014) 111:14852–7. doi: 10.1073/pnas.1403271111
PubMed Abstract | CrossRef Full Text | Google Scholar
31. Chen Ten, Jensen PE. MHC Class Ii Antigen Presentation and Immunological Abnormalities Due to Deficiency of MHC Grade 2 and its Associated Genes. Exp Mol Pathol (2008) 85:twoscore–4. doi: 10.1016/j.yexmp.2008.03.011
PubMed Abstract | CrossRef Full Text | Google Scholar
33. Whitmire JK, Asano MS, Kaech SM, Sarkar S, Hannum LG, Shlomchik MJ, et al. Requirement of B Cells for Generating CD4+ T Cell Retention. J Immunol (2009) 182:1868–76. doi: x.4049/jimmunol.0802501
PubMed Abstract | CrossRef Total Text | Google Scholar
34. Sadegh-Nasseri S, Dalai SK, Korb Ferris LC, Mirshahidi S. Suboptimal Engagement of the T-cell Receptor by a Variety of peptide-MHC Ligands Triggers T-cell Anergy. Immunology (2010) 129:1–seven. doi: 10.1111/j.1365-2567.2009.03206.10
PubMed Abstract | CrossRef Full Text | Google Scholar
35. Misumi I, Whitmire JK. B Cell Depletion Curtails CD4+ T Cell Memory and Reduces Protection Against Disseminating Virus Infection. J Immunol (2014) 192:1597–608. doi: ten.4049/jimmunol.1302661
PubMed Abstract | CrossRef Total Text | Google Scholar
36. Dubois Cauwelaert N, Baldwin SL, Orr MT, Desbien AL, Cuff E, Hofmeyer KA, et al. Antigen Presentation past B Cells Guides Programing of Retention CD4(+) T-Jail cell Responses to a TLR4-agonist Containing Vaccine in Mice. Eur J Immunol (2016) 46:2719–29. doi: 10.1002/eji.201646399
PubMed Abstract | CrossRef Full Text | Google Scholar
38. Korb LC, Mirshahidi S, Ramyar K, Sadighi Akha AA, Sadegh-Nasseri Due south. Induction of T Cell Anergy by Low Numbers of Agonist Ligands. J Immunol (1999) 162:6401–9.
PubMed Abstract | Google Scholar
40. Mirshahidi South, Ferris LC, Sadegh-Nasseri S. The Magnitude of TCR Date is a Critical Predictor of T Jail cell Anergy or Activation. J Immunol (2004) 172:5346–55. doi: 10.4049/jimmunol.172.ix.5346
PubMed Abstruse | CrossRef Full Text | Google Scholar
41. Ryan KR, McNeil LK, Dao C, Jensen PE, Evavold BD. Modification of Peptide Interaction With MHC Creates TCR Partial Agonists. Cell Immunol (2004) 227:lxx–8. doi: ten.1016/j.cellimm.2004.01.003
PubMed Abstract | CrossRef Full Text | Google Scholar
42. Ford ML, Evavold BD. Regulation of Polyclonal T Cell Responses by an MHC Ballast-Substituted Variant of Myelin Oligodendrocyte Glycoprotein 35-55. J Immunol (2003) 171:1247–54. doi: 10.4049/jimmunol.171.3.1247
PubMed Abstruse | CrossRef Full Text | Google Scholar
43. Robertson JM, Evavold BD. Cut Edge: Dueling TCRs: Peptide Animosity of CD4+ T Cells With Dual Antigen Specificities. J Immunol (1999) 163:1750–4.
PubMed Abstract | Google Scholar
44. Mirshahidi S, Huang CT, Sadegh-Nasseri Due south. Anergy in Peripheral Memory CD4(+) T Cells Induced by Low Avidity Engagement of T Cell Receptor. J Exp Med (2001) 194:719–31. doi: 10.1084/jem.194.6.719
PubMed Abstract | CrossRef Full Text | Google Scholar
45. Garside P, Ingulli E, Merica RR, Johnson JG, Noelle RJ, Jenkins MK. Visualization of Specific B and T Lymphocyte Interactions in the Lymph Node. Science (1998) 281:96–9. doi: ten.1126/science.281.5373.96
PubMed Abstract | CrossRef Full Text | Google Scholar
46. Vocal Due north, Sengupta S, Khoruzhenko South, Welsh RA, Kim A, Kumar MR, et al. Multiple Genetic Programs Contribute to CD4 T Cell Memory Differentiation and Longevity by Maintaining T Jail cell Quiescence. Cell Immunol (2020) 357:104210. doi: x.1016/j.cellimm.2020.104210
PubMed Abstruse | CrossRef Total Text | Google Scholar
47. Tokoyoda K, Zehentmeier S, Hegazy AN, Albrecht I, Grun JR, Lohning G, et al. Professional Memory CD4+ T Lymphocytes Preferentially Reside and Rest in the Bone Marrow. Immunity (2009) xxx:721–xxx. doi: ten.1016/j.immuni.2009.03.015
PubMed Abstract | CrossRef Total Text | Google Scholar
49. Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, et al. Enhancing CD8 T-Cell Memory by Modulating Fatty Acid Metabolism. Nature (2009) 460:103–7. doi: 10.1038/nature08097
PubMed Abstruse | CrossRef Full Text | Google Scholar
53. Kim C, Jay DC, Williams MA. Dynamic Functional Modulation of CD4+ T Prison cell Recall Responses is Dependent on the Inflammatory Environment of the Secondary Stimulus. PLoS Pathog (2014) x:e1004137. doi: 10.1371/journal.ppat.1004137
PubMed Abstract | CrossRef Full Text | Google Scholar
54. Welsh RA, Song North, Foss CA, Boronina T, Cole RN, Sadegh-Nasseri Due south. Lack of the MHC Class Ii Chaperone H2-O Causes Susceptibility to Autoimmune Diseases. PLoS Biol (2020) 18:e3000590. doi: 10.1371/journal.pbio.3000590
PubMed Abstract | CrossRef Full Text | Google Scholar
56. Welsh RA, Sadegh-Nasseri S. The Love and Hate Relationship of HLA-DM/DO in the Pick of Immunodominant Epitopes. Curr Opin Immunol (2020) 64:117–23. doi: 10.1016/j.coi.2020.05.007
PubMed Abstruse | CrossRef Total Text | Google Scholar
57. Sercarz EE, Lehmann PV, Ametani A, Benichou Thou, Miller A, Moudgil K. Dominance and Crypticity of T Cell Antigenic Determinants. Annu Rev Immunol (1993) 11:729–66. doi: ten.1146/annurev.iy.eleven.040193.003501
PubMed Abstract | CrossRef Full Text | Google Scholar
58. Natarajan SK, Assadi K, Sadegh-Nasseri S. Stable Peptide Bounden to MHC Form 2 Molecule is Rapid and is Determined past a Receptive Conformation Shaped past Prior Association With Low Affinity Peptides. J Immunol (1999) 162:4030–6.
PubMed Abstruse | Google Scholar
59. Sadegh-Nasseri Southward, Stern LJ, Wiley DC, Germain RN. MHC Class Two Role Preserved by Low-Affinity Peptide Interactions Preceding Stable Binding. Nature (1994) 370:647–50. doi: 10.1038/370647a0
PubMed Abstract | CrossRef Full Text | Google Scholar
lx. Rabinowitz JD, Vrljic M, Kasson PM, Liang MN, Busch R, Boniface JJ, et al. Formation of a Highly Peptide-Receptive State of Course Two Mhc. Immunity (1998) nine:699–709. doi: 10.1016/S1074-7613(00)80667-six
PubMed Abstract | CrossRef Total Text | Google Scholar
62. Kim A, Hartman IZ, Poore B, Boronina T, Cole RN, Vocal N, et al. Divergent Paths for the Selection of Immunodominant Epitopes From Singled-out Antigenic Sources. Nat Commun (2014) five:5369. doi: 10.1038/ncomms6369
PubMed Abstract | CrossRef Full Text | Google Scholar
64. Zarutskie JA, Busch R, Zavala-Ruiz Z, Rushe M, Mellins ED, Stern LJ. The Kinetic Basis of Peptide Exchange Catalysis by HLA-DM. Proc Natl Acad Sci Usa (2001) 98:12450–5. doi: 10.1073/pnas.211439398
PubMed Abstruse | CrossRef Total Text | Google Scholar
65. Belmares MP, Busch R, Mellins ED, McConnell HM. Formation of Two Peptide/MHC 2 Isomers is Catalyzed Differentially past HLA-DM. Biochemistry (2003) 42:838–47. doi: 10.1021/bi020466p
PubMed Abstract | CrossRef Full Text | Google Scholar
66. Pashine A, Busch R, Belmares MP, Munning JN, Doebele RC, Buckingham M, et al. Interaction of HLA-DR With an Acidic Face of HLA-DM Disrupts Sequence-Dependent Interactions With Peptides. Immunity (2003) 19:183–92. doi: 10.1016/S1074-7613(03)00200-0
PubMed Abstract | CrossRef Full Text | Google Scholar
67. Stratikos Eastward, Wiley DC, Stern LJ. Enhanced Catalytic Action of HLA-DM on the Commutation of Peptides Lacking Backbone Hydrogen Bonds Between Their Due north-terminal Region and the MHC Class 2 Alpha-Chain. J Immunol (2004) 172:1109–17. doi: 10.4049/jimmunol.172.two.1109
PubMed Abstract | CrossRef Full Text | Google Scholar
68. Nicholson MJ, Moradi B, Seth NP, Xing X, Cuny GD, Stein RL, et al. Small Molecules That Enhance the Catalytic Efficiency of HLA-DM. J Immunol (2006) 176:4208–xx. doi: 10.4049/jimmunol.176.7.4208
PubMed Abstract | CrossRef Full Text | Google Scholar
69. Chou CL, Mirshahidi Southward, Su KW, Kim A, Narayan Thousand, Khoruzhenko Southward, et al. Short Peptide Sequences Mimic HLA-DM Functions. Mol Immunol (2008) 45:1935–43. doi: 10.1016/j.molimm.2007.ten.033
PubMed Abstract | CrossRef Full Text | Google Scholar
70. Narayan K, Su KW, Chou CL, Khoruzhenko S, Sadegh-Nasseri S. Hla-DM Mediates Peptide Exchange by Interacting Transiently and Repeatedly With HLA-DR1. Mol Immunol (2009) 46:3157–62. doi: 10.1016/j.molimm.2009.07.001
PubMed Abstract | CrossRef Total Text | Google Scholar
71. Zhou Z, Callaway KA, Weber DA, Jensen PE. Cut Border: HLA-DM Functions Through a Mechanism That Does Non Require Specific Conserved Hydrogen Bonds in Class II MHC-Peptide Complexes. J Immunol (2009) 183:4187–91. doi: 10.4049/jimmunol.0901663
PubMed Abstract | CrossRef Full Text | Google Scholar
72. Anders AK, Call MJ, Schulze MS, Fowler KD, Schubert DA, Seth NP, et al. Hla-DM Captures Partially Empty HLA-DR Molecules for Catalyzed Removal of Peptide. Nat Immunol (2011) 12:54–61. doi: 10.1038/ni.1967
PubMed Abstract | CrossRef Full Text | Google Scholar
73. Painter CA, Negroni MP, Kellersberger KA, Zavala-Ruiz Z, Evans JE, Stern LJ. Conformational Lability in the Grade II MHC 310 Helix and Adjacent Extended Strand Dictate HLA-DM Susceptibility and Peptide Commutation. Proc Natl Acad Sci U Due south A (2011) 108:19329–34. doi: 10.1073/pnas.1108074108
PubMed Abstruse | CrossRef Full Text | Google Scholar
74. Pos W, Sethi DK, Call MJ, Schulze MS, Anders AK, Pyrdol J, et al. Crystal Structure of the HLA-DM-HLA-DR1 Complex Defines Mechanisms for Rapid Peptide Pick. Cell (2012) 151:1557–68. doi: x.1016/j.cell.2012.11.025
PubMed Abstract | CrossRef Total Text | Google Scholar
75. Ferrante A, Templeton M, Hoffman M, Castellini MJ. The Thermodynamic Mechanism of Peptide-MHC Class II Complex Formation is a Determinant of Susceptibility to HLA-DM. J Immunol (2015) 195:1251–61. doi: ten.4049/jimmunol.1402367
PubMed Abstract | CrossRef Full Text | Google Scholar
76. Poluektov YO, Kim A, Hartman IZ, Sadegh-Nasseri Southward. Hla-DO equally the Optimizer of Epitope Selection for MHC Course II Antigen Presentation. PLoS 1 (2013) 8:e71228. doi: 10.1371/journal.pone.0071228
PubMed Abstract | CrossRef Total Text | Google Scholar
78. Nanaware PP, Jurewicz MM, Leszyk J, Shaffer SA, Stern LJ. Hla-DO Modulates the Diversity of the MHC-2 Self-Peptidome. Mol Cell Proteomics (2018) 18(iii):490–503 doi: 10.1074/mcp.RA118.000956
PubMed Abstract | CrossRef Full Text | Google Scholar
79. Yoon T, Macmillan H, Mortimer SE, Jiang Westward, Rinderknecht CH, Stern LJ, et al. Mapping the HLA-Exercise/HLA-DM Circuitous by FRET and Mutagenesis. Proc Natl Acad Sci Us (2012) 109:11276–81. doi: 10.1073/pnas.1113966109
PubMed Abstract | CrossRef Full Text | Google Scholar
80. Guce AI, Mortimer SE, Yoon T, Painter CA, Jiang W, Mellins ED, et al. Hla-DO Acts as a Substrate Mimic to Inhibit HLA-DM by a Competitive Mechanism. Nat Struct Mol Biol (2013) 20:90–viii. doi: 10.1038/nsmb.2460
PubMed Abstract | CrossRef Total Text | Google Scholar
81. Kropshofer H, Vogt AB, Thery C, Armandola EA, Li BC, Moldenhauer G, et al. A Role for HLA-DO every bit a Co-Chaperone of HLA-DM in Peptide Loading of MHC Class II Molecules. EMBO J (1998) 17:2971–81. doi: x.1093/emboj/17.eleven.2971
PubMed Abstract | CrossRef Full Text | Google Scholar
84. Roucard C, Thomas C, Pasquier MA, Trowsdale J, Sotto JJ, Neefjes J, et al. In Vivo and In Vitro Modulation of HLA-DM and HLA-Do is Induced by B Lymphocyte Activation. J Immunol (2001) 167:6849–58. doi: 10.4049/jimmunol.167.12.6849
PubMed Abstract | CrossRef Full Text | Google Scholar
85. Yeh CH, Nojima T, Kuraoka One thousand, Kelsoe G. Germinal Center Entry Not Selection of B Cells is Controlled by peptide-MHCII Complex Density. Nat Commun (2018) 9:928. doi: ten.1038/s41467-018-03382-ten
PubMed Abstruse | CrossRef Full Text | Google Scholar
86. Mesin Fifty, Schiepers A, Ersching J, Barbulescu A, Cavazzoni CB, Angelini A, et al. Restricted Clonality and Limited Germinal Center Reentry Characterize Retentiveness B Cell Reactivation by Boosting. Cell (2020) 180:92–106.e11. doi: 10.1016/j.jail cell.2019.11.032
PubMed Abstruse | CrossRef Full Text | Google Scholar
87. Bannard O, McGowan SJ, Ersching J, Ishido Southward, Victora GD, Shin JS, et al. Ubiquitin-Mediated Fluctuations in MHC Course II Facilitate Efficient Germinal Center B Prison cell Responses. J Exp Med (2016) 213:993–1009. doi: ten.1084/jem.20151682
PubMed Abstract | CrossRef Full Text | Google Scholar
88. Shulman Z, Gitlin Ad, Weinstein JS, Lainez B, Esplugues East, Flavell RA, et al. Dynamic Signaling by T Follicular Helper Cells During Germinal Centre B Jail cell Pick. Science (2014) 345:1058–62. doi: 10.1126/scientific discipline.1257861
PubMed Abstract | CrossRef Full Text | Google Scholar
90. Chalouni C, Banchereau J, Vogt AB, Pascual 5, Davoust J. Human Germinal Centre B Cells Differ From Naive and Memory B Cells past Their Aggregated MHC Class 2-Rich Compartments Defective HLA-practise. Int Immunol (2003) 15:457–66. doi: 10.1093/intimm/dxg037
PubMed Abstruse | CrossRef Full Text | Google Scholar
91. Glazier KS, Hake SB, Tobin HM, Chadburn A, Schattner EJ, Denzin LK. Germinal Centre B Cells Regulate Their Adequacy to Nowadays Antigen by Modulation of HLA-DO. J Exp Med (2002) 195:1063–9. doi: 10.1084/jem.20012059
PubMed Abstract | CrossRef Full Text | Google Scholar
92. Chen 10, Laur O, Kambayashi T, Li S, Bray RA, Weber DA, et al. Regulated Expression of Man Histocompatibility Leukocyte Antigen (HLA)-DO During Antigen-Dependent and Antigen-Contained Phases of B Prison cell Development. J Exp Med (2002) 195:1053–62. doi: 10.1084/jem.20012066
PubMed Abstruse | CrossRef Full Text | Google Scholar
93. Janssen EM, Lemmens EE, Wolfe T, Christen U, von Herrath MG, Schoenberger SP. Cd4+ T Cells Are Required for Secondary Expansion and Memory in CD8+ T Lymphocytes. Nature (2003) 421:852–vi. doi: 10.1038/nature01441
PubMed Abstract | CrossRef Full Text | Google Scholar
Source: https://www.frontiersin.org/articles/10.3389/fimmu.2021.677036/full
0 Response to "Review of B Cells Cd4+ T Cells and Cd8+ T Cells"
Post a Comment